Most Important Biology Diagram For NEET UG 2026: Welcome to Day 8 of our Most Important Biology Diagram series- where complex NCERT concepts turn into crystal-clear visuals you’ll never forget. Because let’s be honest, one well-understood diagram can save you from rereading ten pages of theory! So grab your notebook, sharpen that pencil, and let’s decode today’s high-yield diagram that could quietly add a few extra marks to your NEET score.
Let’s start today’s practice session:
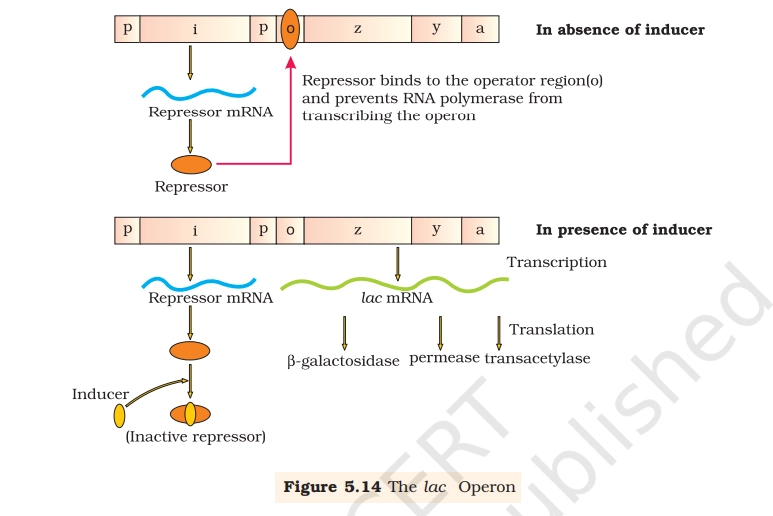
Lac Operon Diagram
The lac operon is a genetic regulatory system in E. coli that controls lactose metabolism genes, turning them on only when lactose is present and glucose is absent to conserve energy.
Core Components
- Regulatory gene (i) → produces repressor protein continuously (constitutive expression).
- Promoter (p) → binding site for RNA polymerase.
- Operator (o) → binding site for repressor; acts as the switch.
- Structural genes:
- z → β-galactosidase
- y → permease
- a → transacetylase
In Absence of Inducer (Lactose)
- Repressor protein binds to operator.
- RNA polymerase cannot transcribe structural genes.
- No enzyme production → operon is OFF.
In Presence of Inducer (Lactose)
- Lactose (actually allolactose) acts as inducer.
- Inducer binds repressor → inactivates it.
- Repressor cannot bind operator.
- RNA polymerase transcribes z, y, a genes → enzymes formed.
- Operon becomes ON (inducible system).
Extra NEET-Level Facts
- Lac operon is an example of negative regulation.
- Shows inducible operon (normally OFF, turned ON by substrate).
- Permease allows lactose entry; β-galactosidase breaks lactose → glucose + galactose.
- Glucose presence suppresses lac operon (catabolite repression via cAMP-CAP).
PRACTICE QUESTIONS
Q.1. In the lac operon of E. coli, which correctly matches each structural gene with its primary product?
A. lacZ – permease, lacY – β-galactosidase, lacA – transacetylase
B. lacZ – transacetylase, lacY – permease, lacA – β-galactosidase
C. lacZ – β-galactosidase, lacY – permease, lacA – transacetylase
D. lacZ – β-galactosidase, lacY – transacetylase, lacA – permease
Q.2. In a standard diagram of the lac operon, which sequence correctly represents the order of elements along the DNA from the regulatory gene towards the structural genes?
A. lacI – operator (O) – promoter (P) – lacZ – lacY – lacA
B. Operator (O) – lacI – promoter (P) – lacZ – lacY – lacA
C. lacI – promoter (P) – operator (O) – lacZ – lacY – lacA
D. Promoter (P) – lacI – operator (O) – lacZ – lacY – lacA
Q.3. Which statement best explains the role of the lac repressor protein in the regulation of the lac operon?
A. It forms cyclic AMP, which then activates transcription of the operon
B. It catalyzes the breakdown of lactose into glucose and galactose
C. It binds directly to the promoter to recruit RNA polymerase when lactose is present
D. It binds to the operator to block transcription when lactose is absent
Q.4. In which condition will the lac operon show maximal transcription of its structural genes?
A. Low glucose and high lactose in the medium
B. High glucose and high lactose in the medium
C. High glucose and no lactose in the medium
D. No glucose and no lactose in the medium
Q.5. What immediate molecular change occurs when allolactose binds to the lac repressor protein?
A. The repressor undergoes a conformational change and can no longer bind the operator
B. The repressor dimerizes and binds more strongly to the operator
C. The repressor is degraded by proteases, removing it from the cell
D. The repressor begins to synthesize cAMP, activating CAP
Read Also: Most Important Biology Diagram For NEET UG 2026 (Day 7)
Process of Transcription in Bacteria
Transcription is the process by which genetic information from DNA is copied into RNA by the enzyme RNA polymerase, using one DNA strand as a template.
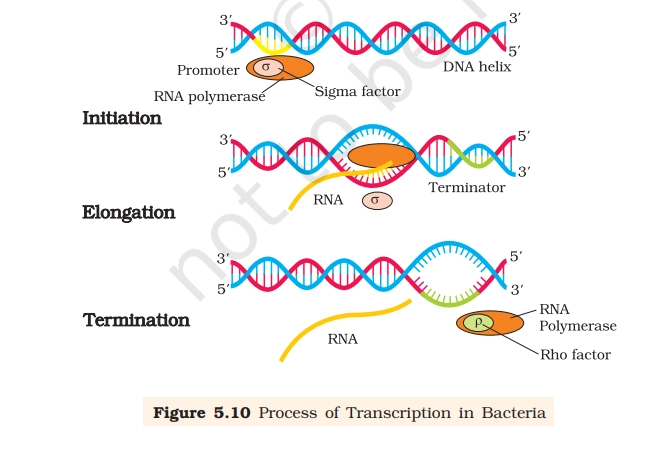
1. Initiation
- RNA polymerase + sigma (σ) factor forms holoenzyme.
- Holoenzyme recognizes promoter region on DNA.
- DNA unwinds locally → transcription bubble formed.
- First phosphodiester bond is formed using nucleoside triphosphates (NTPs).
- After initiation, σ-factor dissociates.
2. Elongation
- Core RNA polymerase moves along template strand (3′ → 5′).
- RNA is synthesized in 5′ → 3′ direction.
- Complementary base pairing followed:
- A-U, G-C.
- Only a small stretch of RNA remains attached to DNA inside enzyme.
- DNA behind polymerase re-anneals.
3. Termination
- When polymerase reaches terminator sequence, transcription stops.
- ρ (rho) factor–dependent termination shown in diagram.
- RNA transcript released, polymerase detaches, DNA helix reforms.
4. Extra NEET Facts
- In bacteria, transcription and translation are coupled (occur simultaneously).
- Single RNA polymerase synthesizes all types of RNA in prokaryotes.
- No mRNA processing (no capping, splicing, tailing) in bacteria.
PRACTICE QUESTIONS
Q.6. In a typical bacterial promoter, where does the sigma factor of RNA polymerase holoenzyme primarily recognize and bind to initiate transcription?
A. Directly to the +1 nucleotide where transcription begins
B. To the conserved -10 and -35 regions upstream of the transcription start site
C. Within the coding region just downstream of the start codon
D. To the terminator region to position RNA polymerase correctly
Q.7. Which statement best describes the role of sigma factor in the bacterial transcription initiation process?
A. It forms phosphodiester bonds between ribonucleotides during elongation
B. It splices introns out of the pre-mRNA transcript
C. It recognizes promoter sequences and then dissociates after initiation
D. It terminates transcription by causing RNA polymerase to fall off DNA
Q.8. In a diagram of bacterial transcription, which region would you label as the “promoter”?
A. The DNA sequence encoding the amino acid sequence of the protein
B. The DNA sequence where RNA polymerase first binds with the help of sigma factor
C. The RNA sequence rich in G and C that forms a hairpin
D. The protein factor that binds at the termination site
Q.9. During bacterial transcription, what happens immediately after RNA polymerase holoenzyme successfully initiates RNA synthesis and clears the promoter?
A. Sigma factor remains tightly bound and guides elongation until termination
B. Sigma factor dissociates, and the core polymerase continues elongation
C. The DNA promoter sequence is removed from the chromosome
D. Rho factor binds to the promoter and blocks further transcription
Q.10. Which event directly triggers rho-dependent termination of transcription in bacteria?
A. Sigma factor rejoining the core enzyme at the terminator
B. Formation of a G-C-rich hairpin in the RNA followed by a U-rich region
C. Binding of rho factor to the promoter at the -35 region
D. Rho factor catching up with RNA polymerase at a pause site and causing release
Comment Below for Answers
Read Also: NEET UG 2026: Daily Practice Questions To Score 650+ (DAY 22)

